clustOpt: Quickstart Guide
Compiled: 2025-08-01
Source:vignettes/quick_start_guide.Rmd
quick_start_guide.Rmd
library(Seurat)
# Only v5 Seurat Assays are supported
options(Seurat.object.assay.version = "v5")
# Increase future globals limit for large datasets
options(future.globals.maxSize = 2e9) # 2GB limit
library(dplyr)
library(tidyr)
library(ggplot2)
library(clustOpt)
library(future)
set.seed(42)Read in the tutorial dataset. This data was generated from the Asian Immune Diversity Atlas (data freeze v1). It contains only B cells, Monocytes, and NK cells from 10 donors which have been sketched to a total of 1000 cells using the leverage score based method of Seurat’s SketchData function.
input <- readRDS(
system.file(
"extdata",
"1000_cell_sketch_10_donors_3_celltypes_AIDA.rds",
package = "clustOpt"
)
)
input@meta.data |>
summarize(
Donors = n_distinct(donor_id),
Celltypes = n_distinct(broad_cell_type),
`Cell Subtypes` = n_distinct(author_cell_type),
Ethnicities = n_distinct(Ethnicity_Selfreported),
Countries = n_distinct(Country)
) |>
pivot_longer(cols = everything(), names_to = "Metric", values_to = "Count")
#> # A tibble: 5 × 2
#> Metric Count
#> <chr> <int>
#> 1 Donors 10
#> 2 Celltypes 3
#> 3 Cell Subtypes 10
#> 4 Ethnicities 5
#> 5 Countries 3
input <- input |>
SCTransform(verbose = FALSE) |>
RunPCA(verbose = FALSE)
ElbowPlot(input, ndims = 50)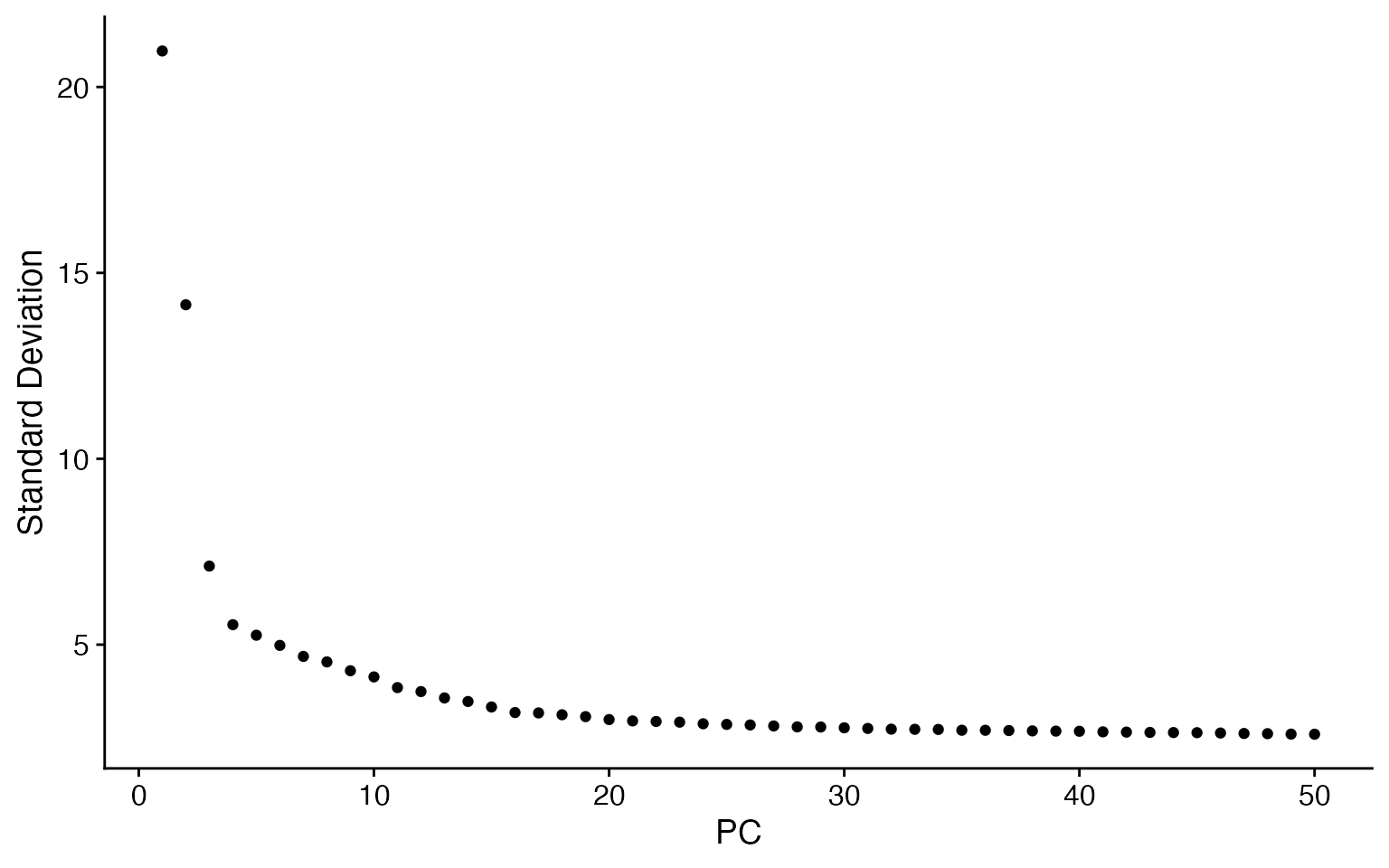
input <- RunUMAP(input, dims = 1:20, verbose = FALSE)
DimPlot(input,
group.by = "donor_id",
pt.size = .8
) +
coord_fixed()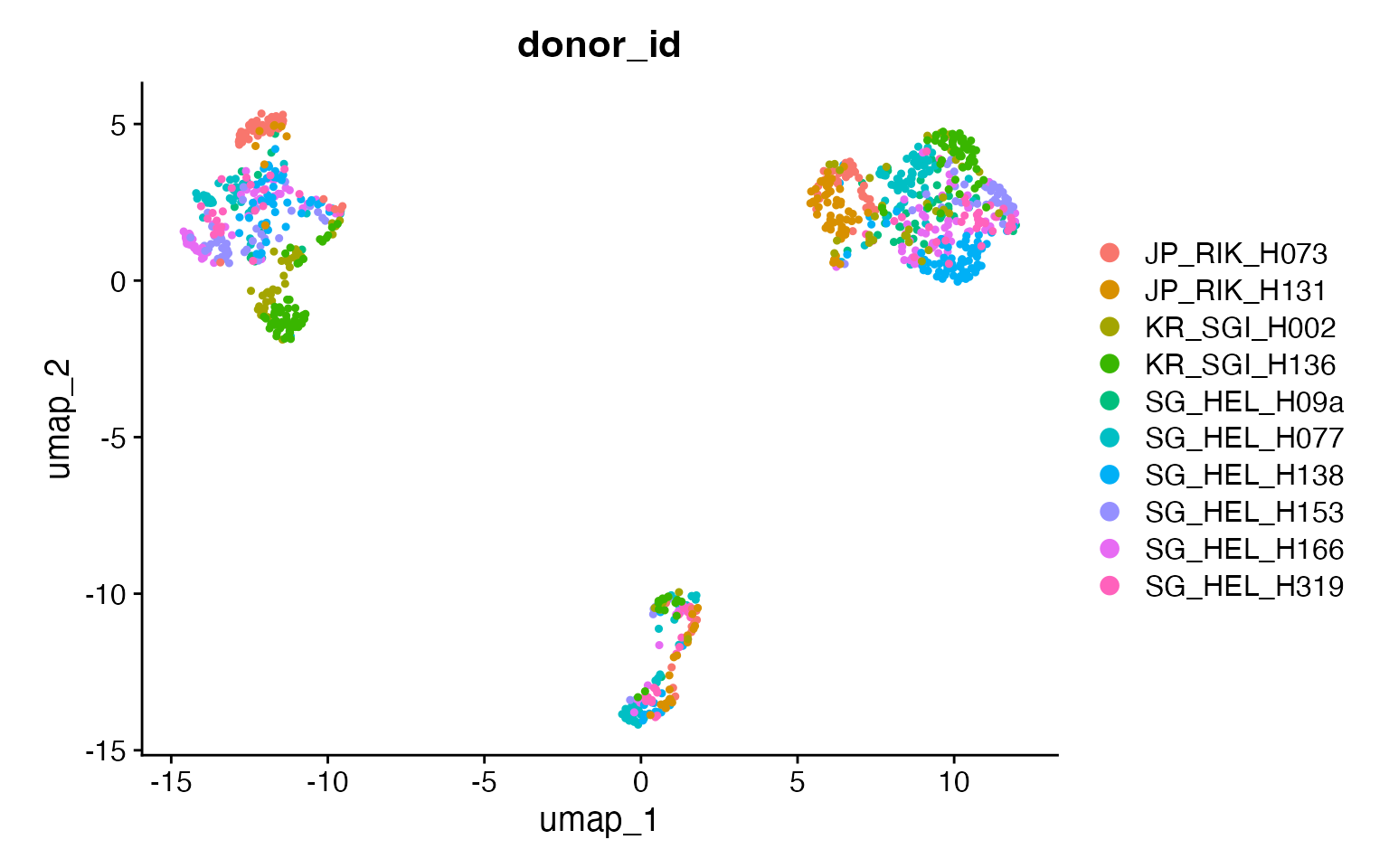
DimPlot(input,
group.by = "broad_cell_type",
pt.size = .8
) +
coord_fixed()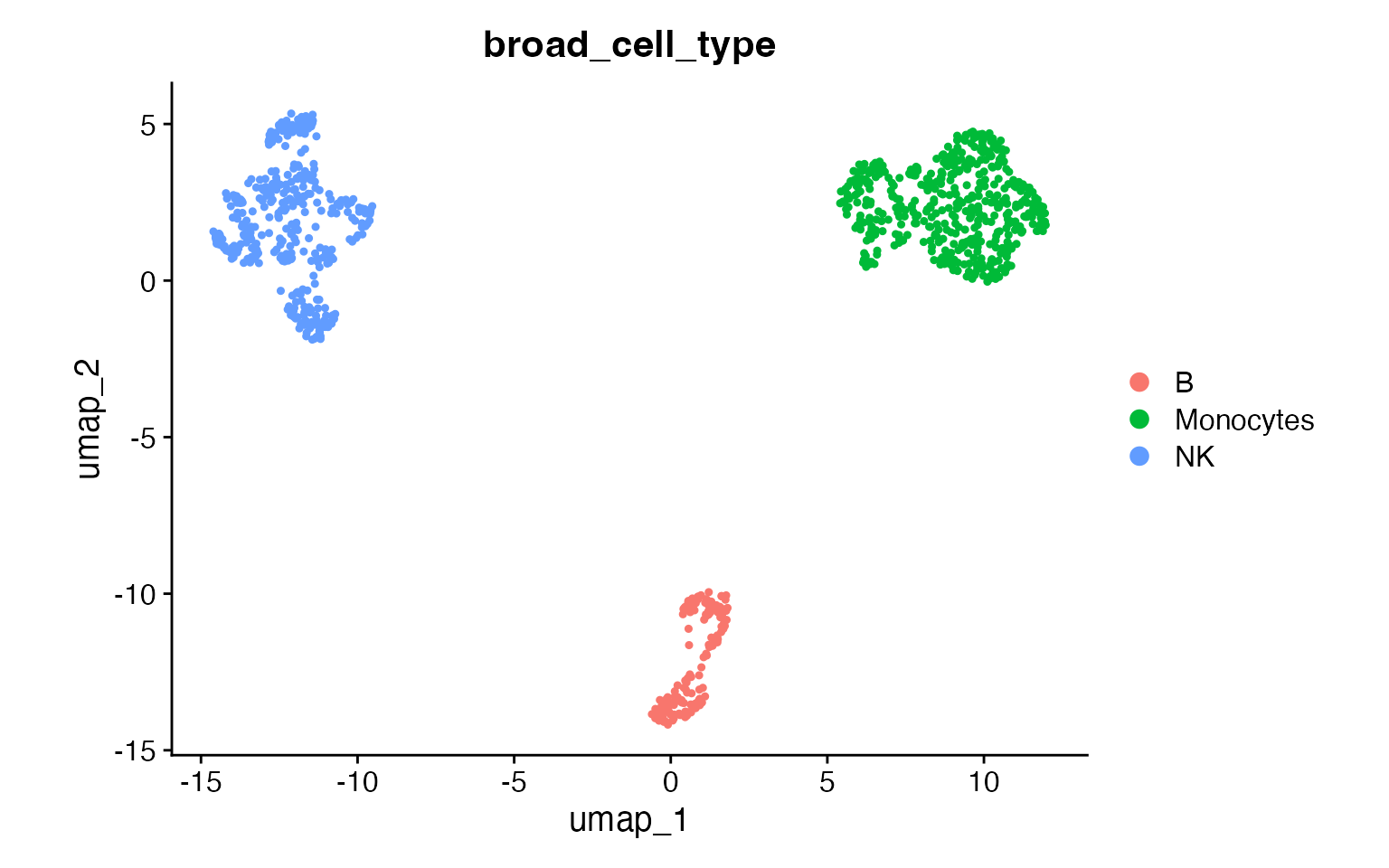
DimPlot(input,
group.by = "author_cell_type",
pt.size = .8
) +
coord_fixed()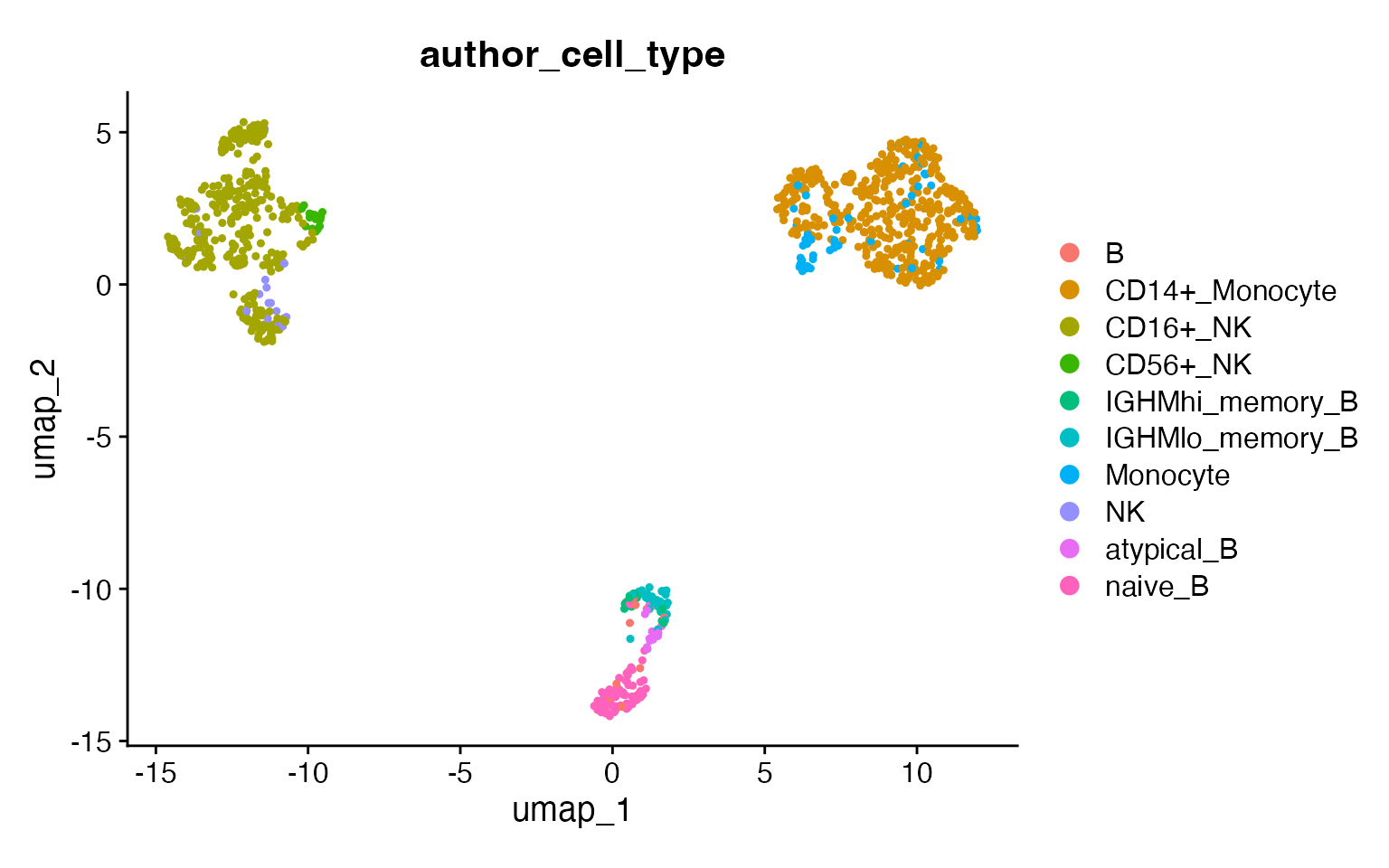
Subject-wise cross validation is parallelized with
future and future.batchtools can be used for
HPC job schedulers. For details on which plan to use for your setup, see
the future package documentation. The code below requires a large amount
of RAM (32 GB recommended) and 10 cores to run in about 20 minutes. The
repeated messages about “SeuratObject” are a side effect of using the
future plan “multisession” and should not occur with other future
plans.
plan("multisession", workers = 10)
sil_dist <- clust_opt(input,
subject_ids = "donor_id",
ndim = 20,
verbose = FALSE,
train_with = "even"
) |>
progressr::with_progress() # Optional, just adds progress bar
#> Input is small enough to run with all cells
#> Using 10 subject(s) that have at least 50 cells:
#> JP_RIK_H073 (100 cells)
#> JP_RIK_H131 (83 cells)
#> KR_SGI_H002 (76 cells)
#> KR_SGI_H136 (127 cells)
#> SG_HEL_H09a (52 cells)
#> SG_HEL_H077 (147 cells)
#> SG_HEL_H138 (130 cells)
#> SG_HEL_H153 (85 cells)
#> SG_HEL_H166 (123 cells)
#> SG_HEL_H319 (77 cells)
#> Found 110 combinations of test subject and resolution
#> Holdout subject: JP_RIK_H073
#> Loading required package: SeuratObject
#> Loading required package: sp
#>
#> Attaching package: ‘SeuratObject’
#> The following objects are masked from ‘package:base’:
#>
#> intersect, t
#> Loading required package: SeuratObject
#> Loading required package: sp
#>
#> Attaching package: ‘SeuratObject’
#> The following objects are masked from ‘package:base’:
#>
#> intersect, t
#> Loading required package: SeuratObject
#> Loading required package: sp
#>
#> Attaching package: ‘SeuratObject’
#> The following objects are masked from ‘package:base’:
#>
#> intersect, t
#> Loading required package: SeuratObject
#> Loading required package: sp
#>
#> Attaching package: ‘SeuratObject’
#> The following objects are masked from ‘package:base’:
#>
#> intersect, t
#> Loading required package: SeuratObject
#> Loading required package: sp
#>
#> Attaching package: ‘SeuratObject’
#> The following objects are masked from ‘package:base’:
#>
#> intersect, t
#> Loading required package: SeuratObject
#> Loading required package: sp
#>
#> Attaching package: ‘SeuratObject’
#> The following objects are masked from ‘package:base’:
#>
#> intersect, t
#> Loading required package: SeuratObject
#> Loading required package: sp
#>
#> Attaching package: ‘SeuratObject’
#> The following objects are masked from ‘package:base’:
#>
#> intersect, t
#> Loading required package: SeuratObject
#> Loading required package: sp
#>
#> Attaching package: ‘SeuratObject’
#> The following objects are masked from ‘package:base’:
#>
#> intersect, t
#> Loading required package: SeuratObject
#> Loading required package: sp
#>
#> Attaching package: ‘SeuratObject’
#> The following objects are masked from ‘package:base’:
#>
#> intersect, t
#> Loading required package: SeuratObject
#> Loading required package: sp
#>
#> Attaching package: ‘SeuratObject’
#> The following objects are masked from ‘package:base’:
#>
#> intersect, t
#> Loading required package: SeuratObject
#> Loading required package: sp
#>
#> Attaching package: ‘SeuratObject’
#> The following objects are masked from ‘package:base’:
#>
#> intersect, t
#> Loading required package: SeuratObject
#> Loading required package: sp
#>
#> Attaching package: ‘SeuratObject’
#> The following objects are masked from ‘package:base’:
#>
#> intersect, t
#> Loading required package: SeuratObject
#> Loading required package: sp
#>
#> Attaching package: ‘SeuratObject’
#> The following objects are masked from ‘package:base’:
#>
#> intersect, t
#> Loading required package: SeuratObject
#> Loading required package: sp
#>
#> Attaching package: ‘SeuratObject’
#> The following objects are masked from ‘package:base’:
#>
#> intersect, t
#> Loading required package: SeuratObject
#> Loading required package: sp
#>
#> Attaching package: ‘SeuratObject’
#> The following objects are masked from ‘package:base’:
#>
#> intersect, t
#> Loading required package: SeuratObject
#> Loading required package: sp
#>
#> Attaching package: ‘SeuratObject’
#> The following objects are masked from ‘package:base’:
#>
#> intersect, t
#> Loading required package: SeuratObject
#> Loading required package: sp
#>
#> Attaching package: ‘SeuratObject’
#> The following objects are masked from ‘package:base’:
#>
#> intersect, t
#> Loading required package: SeuratObject
#> Loading required package: sp
#>
#> Attaching package: ‘SeuratObject’
#> The following objects are masked from ‘package:base’:
#>
#> intersect, t
#> Loading required package: SeuratObject
#> Loading required package: sp
#>
#> Attaching package: ‘SeuratObject’
#> The following objects are masked from ‘package:base’:
#>
#> intersect, t
#> Loading required package: SeuratObject
#> Loading required package: sp
#>
#> Attaching package: ‘SeuratObject’
#> The following objects are masked from ‘package:base’:
#>
#> intersect, t
#> Found 2932 (97.73%) shared genes used for projecting test data
#> Holdout subject: JP_RIK_H131
#> Found 2923 (97.43%) shared genes used for projecting test data
#> Holdout subject: KR_SGI_H002
#> Found 2923 (97.43%) shared genes used for projecting test data
#> Holdout subject: KR_SGI_H136
#> Found 2942 (98.07%) shared genes used for projecting test data
#> Holdout subject: SG_HEL_H09a
#> Found 2820 (94.00%) shared genes used for projecting test data
#> Holdout subject: SG_HEL_H077
#> Found 2988 (99.60%) shared genes used for projecting test data
#> Holdout subject: SG_HEL_H138
#> Found 2956 (98.53%) shared genes used for projecting test data
#> Holdout subject: SG_HEL_H153
#> Found 2922 (97.40%) shared genes used for projecting test data
#> Holdout subject: SG_HEL_H166
#> Found 2956 (98.53%) shared genes used for projecting test data
#> Holdout subject: SG_HEL_H319
#> Found 2896 (96.53%) shared genes used for projecting test data
# Shut down stray workers
plan("sequential")Plotting the silhouette score distributions
plots <- create_sil_plots(sil_dist |> drop_na())
plots[[1]]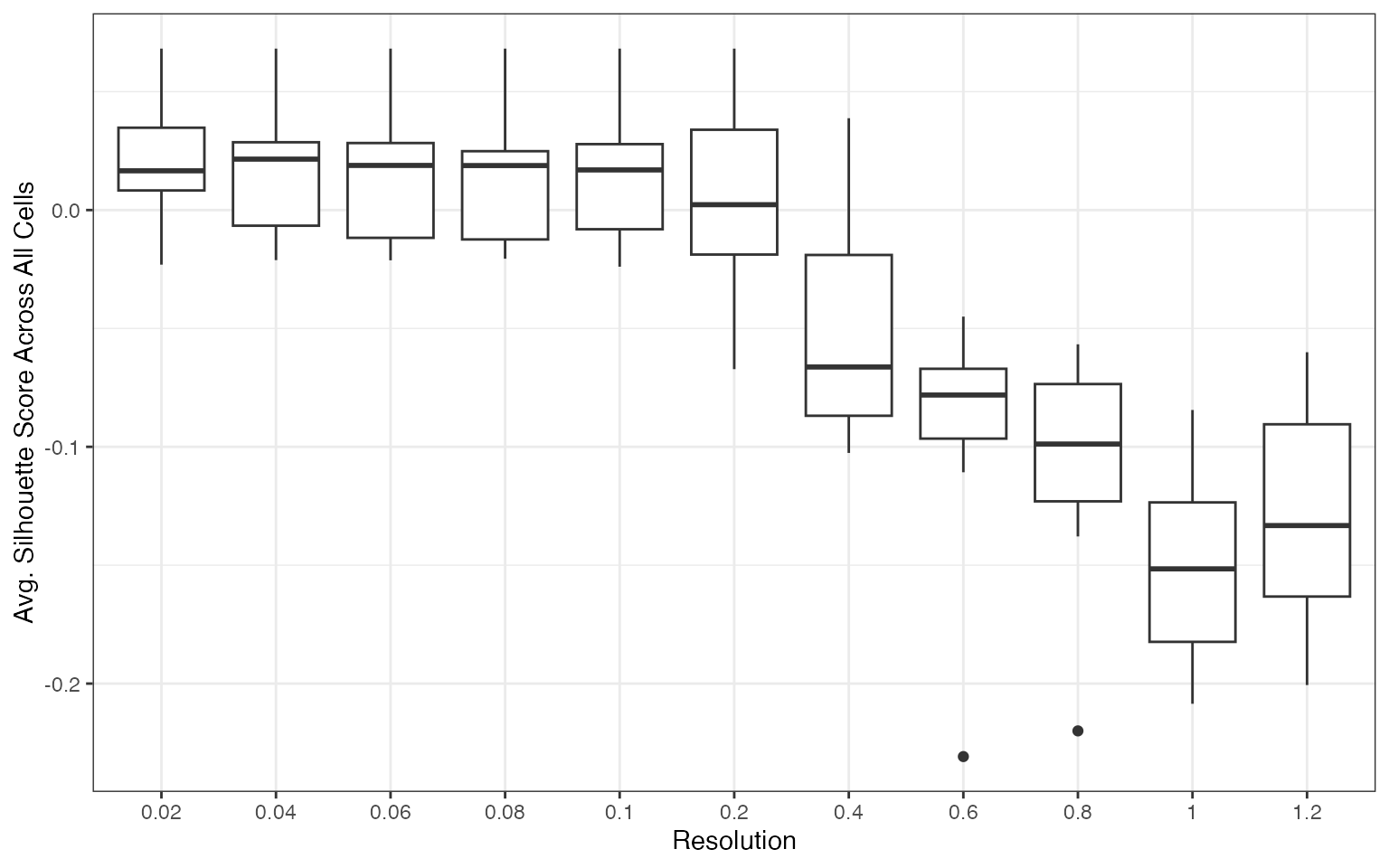
plots[[2]]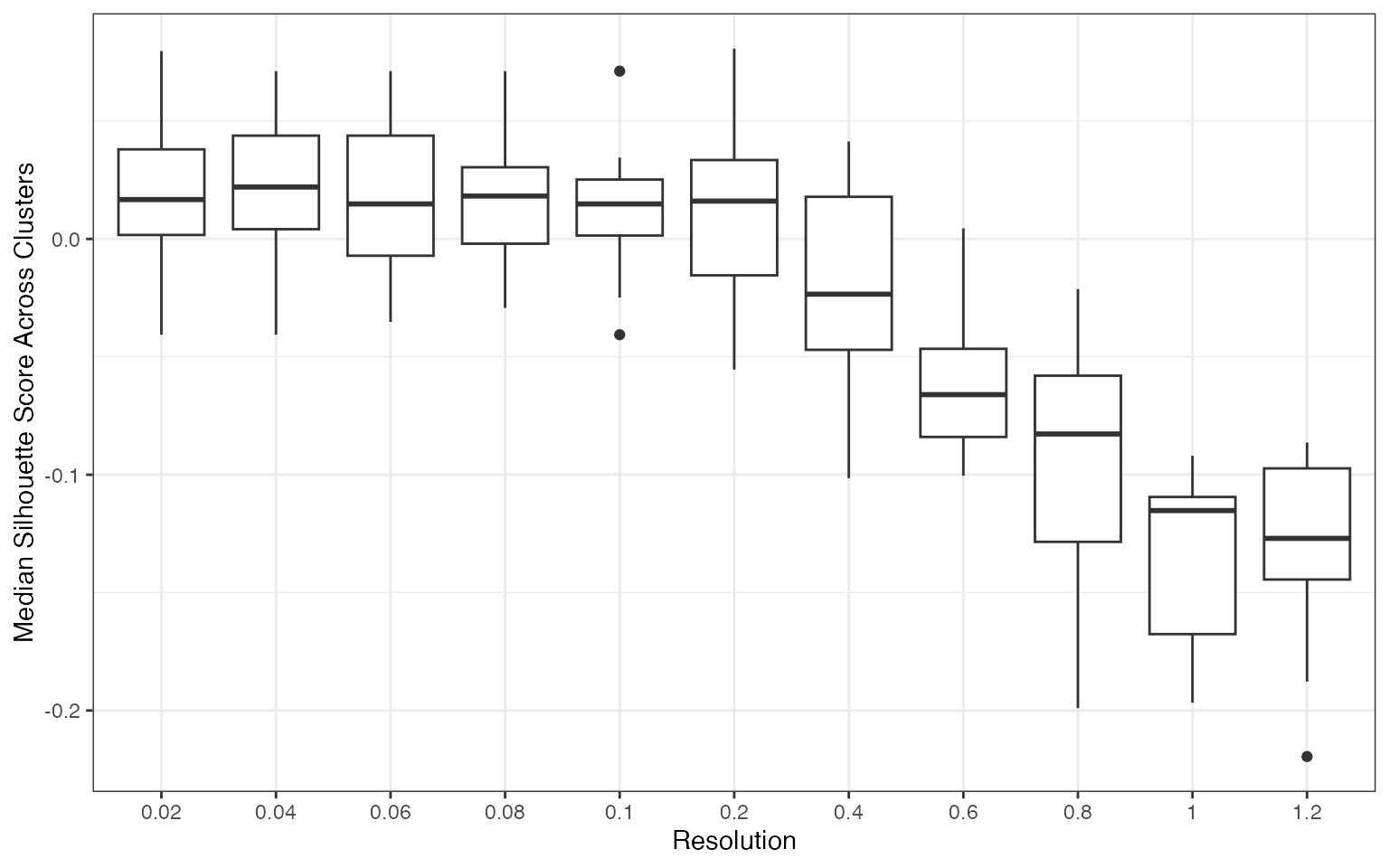
plots[[3]]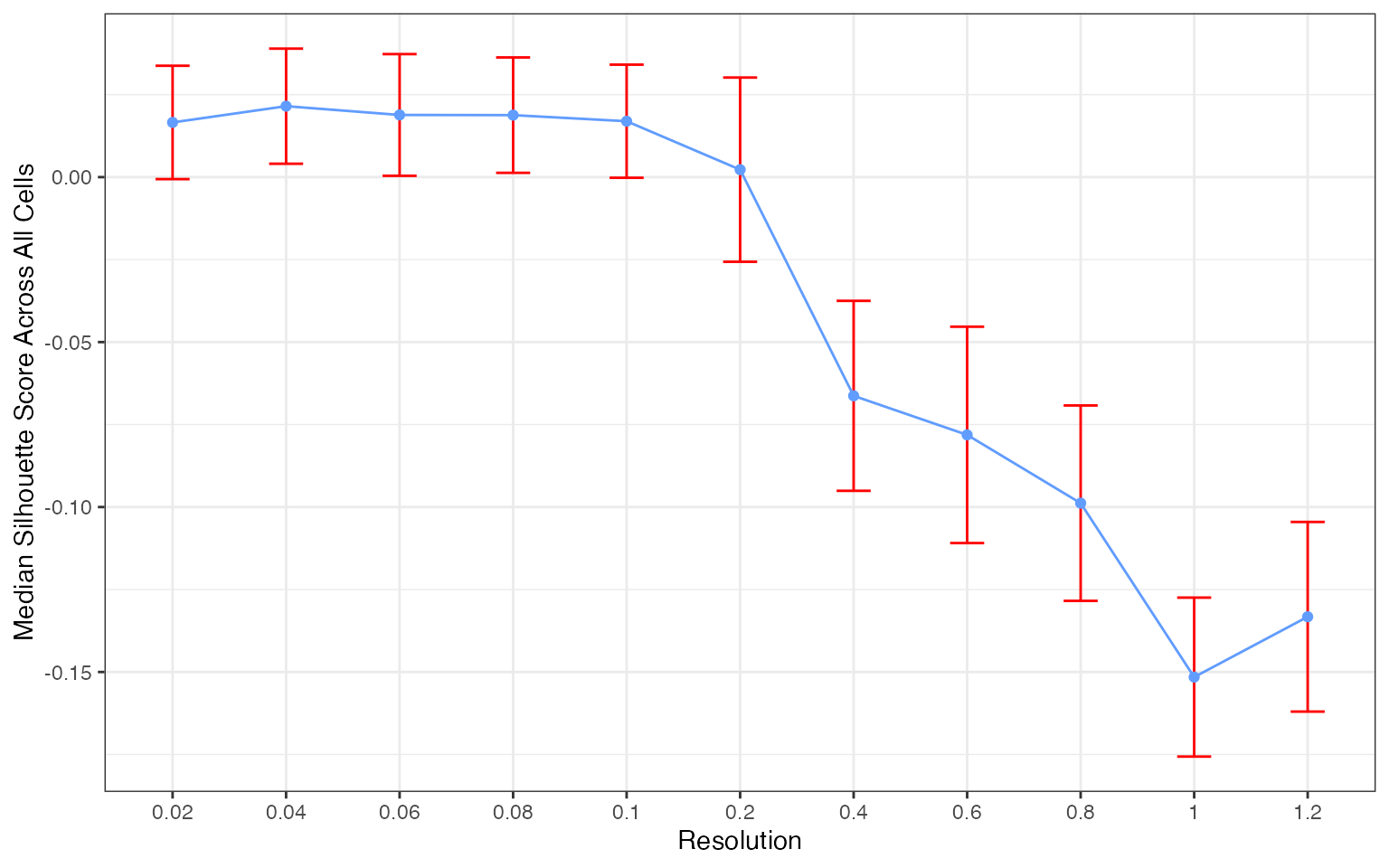
plots[[4]]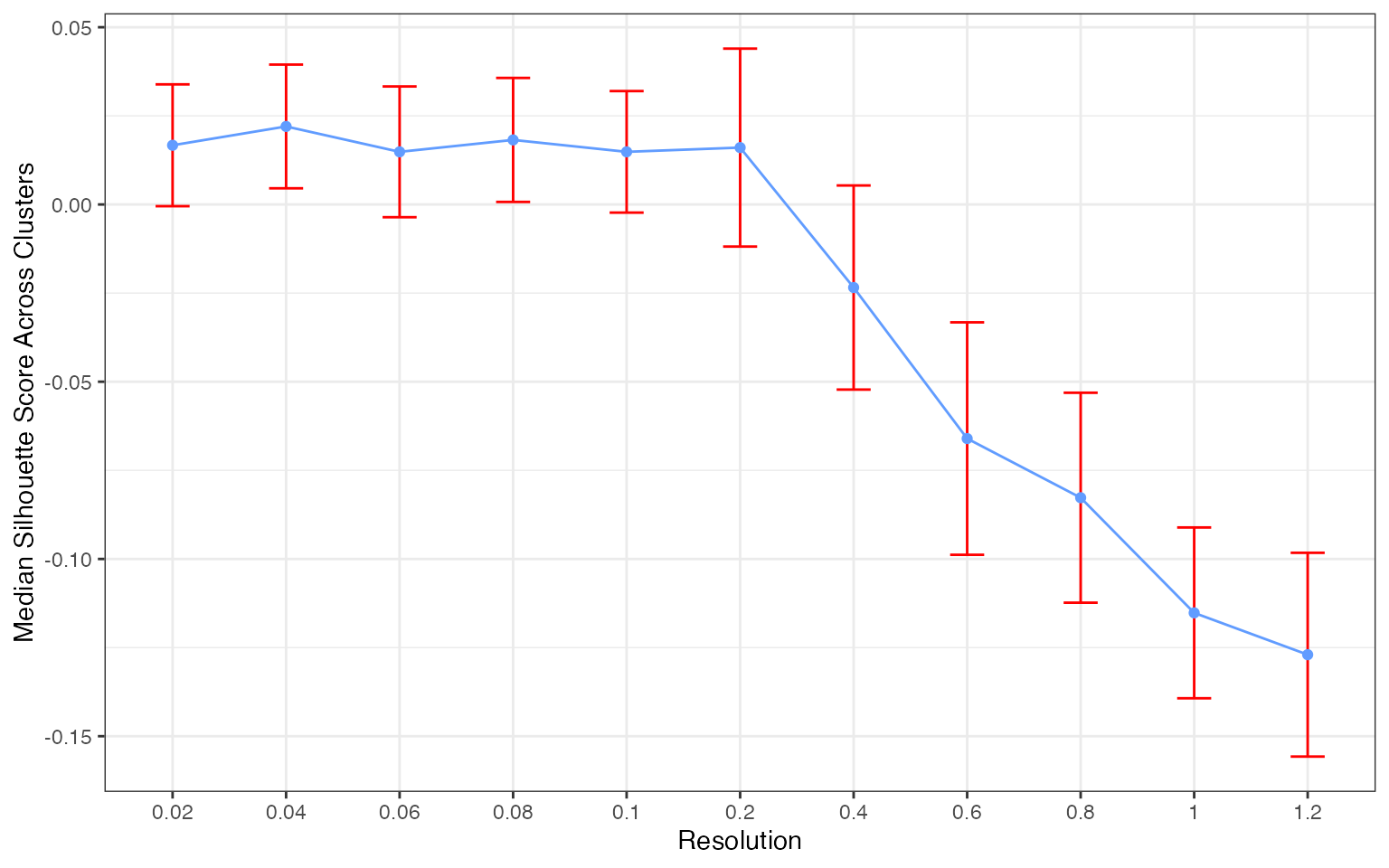
The median silhouette scores across all cells start to fall off at 0.2, indicating that this clustering resolution is the most reproducible across all biological replicates. It is also possible to see a local maximum which would indicate a reproducible resolution.
Comparison to the cell annotations
input <- FindNeighbors(input, dims = 1:20, verbose = FALSE)
input <- FindClusters(input, resolution = 0.2)
#> Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
#>
#> Number of nodes: 1000
#> Number of edges: 31348
#>
#> Running Louvain algorithm...
#> Maximum modularity in 10 random starts: 0.9287
#> Number of communities: 4
#> Elapsed time: 0 seconds
DimPlot(input,
group.by = "seurat_clusters",
pt.size = .8
)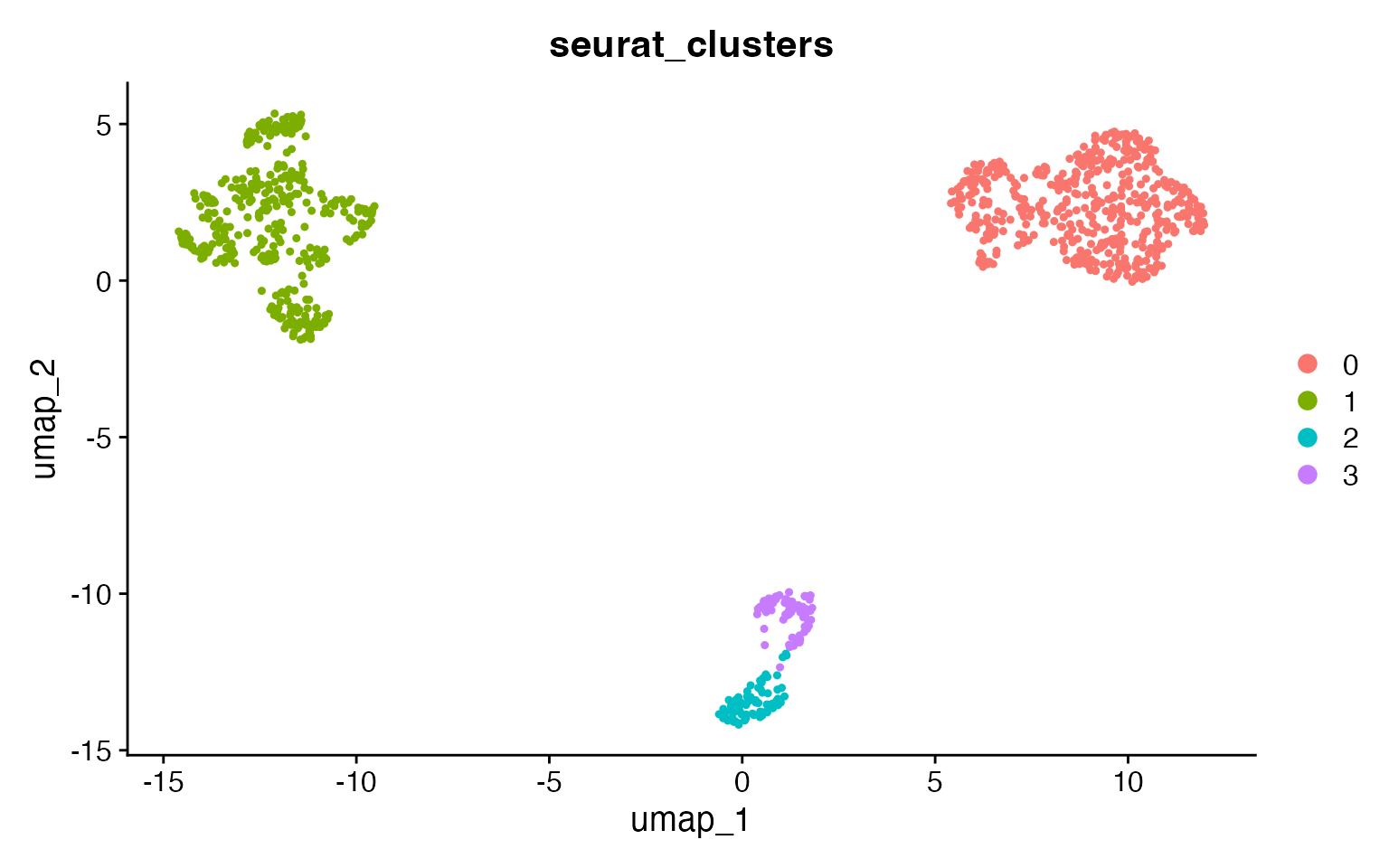
We can see that at the reproducible resolution there are 4 clusters, with the naive B cells being distinct enough to be separated into their own cluster based on the ability to detect them across the CV folds. In other words the naive B cells are present in high enough amounts across the subjects and transcriptomically distinct enough to be consistently detected.
For convenience we provide a function to calculate the adjusted rand index for any 2 metadata columns in a seurat object so that the clustOpt resolution parameter can be compared to clusters derived from other methods.
adjusted_rand_index(input,
meta1 = "seurat_clusters",
meta2 = "broad_cell_type"
)
#> [1] 0.9765531
adjusted_rand_index(input,
meta1 = "seurat_clusters",
meta2 = "author_cell_type"
)
#> [1] 0.8265365Larger Datasets
Due to the resource requirements of clustOpt, we recommend running it
in Rscripts with the future plan set to “multicore” instead
of “multisession” and if possible using future.batchtools.
Additionally we provide a wrapper around Seurat’s
SketchData using the leverage score method to reduce the size of the
input data to clustOpt.
input <- leverage_sketch(input, sketch_size = 100)
#> Sketching scRNA data: 1000 -> 100 cells
#>
#> Removing previously calculated leverage scores...
#> Normalizing scRNA-seq data...
#> Finding variable features for scRNA-seq data...
#> Using 2000 variable features for sketching
#> Calcuating Leverage Score
#> Renaming default assay from sketch to RNA
input
#> An object of class Seurat
#> 13855 features across 100 samples within 1 assay
#> Active assay: RNA (13855 features, 2000 variable features)
#> 3 layers present: counts, data, scale.data