Introduction
Biomedical research very often involves data generated from repeated measures experiments. RMeDPower2 is an R package that provides complete functionality to analyse data coming from repeated measures experiments, i.e., where one has repeated measures from the same biological/independent units or samples.
RMeDPower2 helps test the modeling assumptions one makes, identify outlier observations, outlier units at different levels of the design, estimates statistical power or perform sample size calculations, estimate parameters of interest and also to visualize the association being tested. The functionality is limited to testing associations of one predictor (continuous or categorical, e.g., disease status or brain pathology) along with one another covariate (e.g., gender status) in the context of hierarchical or crossed experimental designs.
RMeDPower2 defines the experimental design for the data, probability model of the data generating distribution and necessary parameters required for sample size calculation using convenient S4 class objects. It uses these objects in one framework that brings together the functionality implemented in multiple R packages - lme4 (implementation of linear mixed effects models), influence.ME (identification of outlier units), EnvStats EnvStats (identification of outlier observations), DHARMa (testing of modeling assumptions for non-normal distributions), simr (sample-size calculations) and tidyverse (data manipulation and visualization).
Installation
install.packages("devtools")
library(devtools)
install_github('gladstone-institutes/RMeDPower2', build_vignettes=TRUE)
library(RMeDPower2)Application Domains
RMeDPower2 supports a wide range of biomedical research applications across different experimental domains:
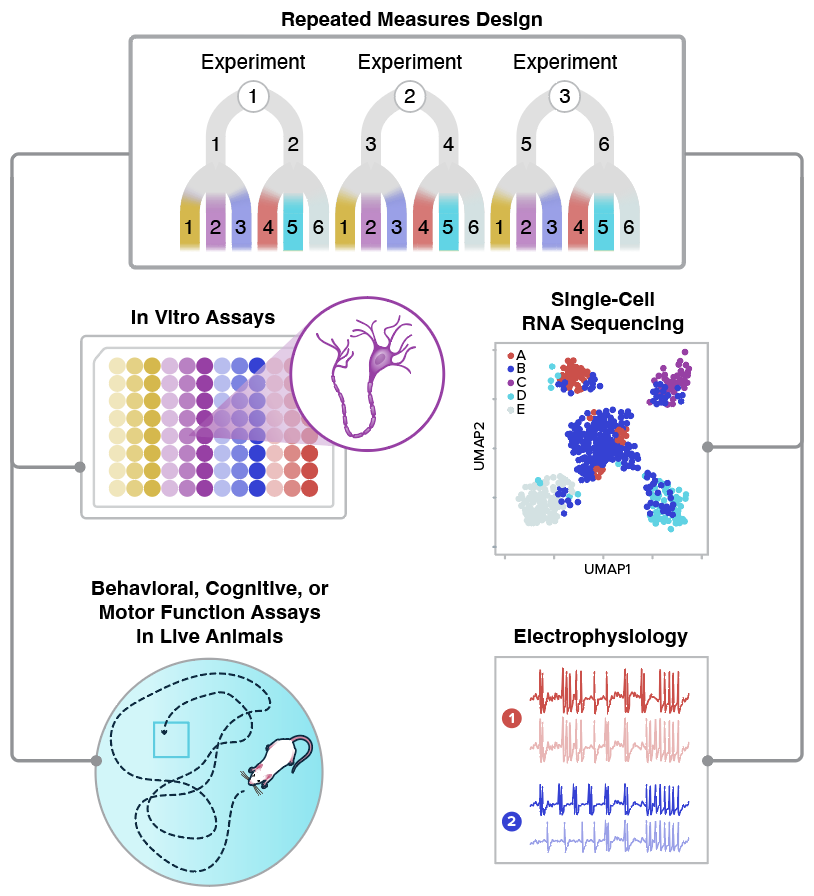
RMeDPower2 can be applied to various biomedical research contexts including cell assays, single-cell RNA-seq, behavioral studies, and electrophysiology experiments. Each domain involves repeated measure experimental designs with nested or crossed factors.
Package Structure
The package is organized around three core S4 classes that define the experimental framework:
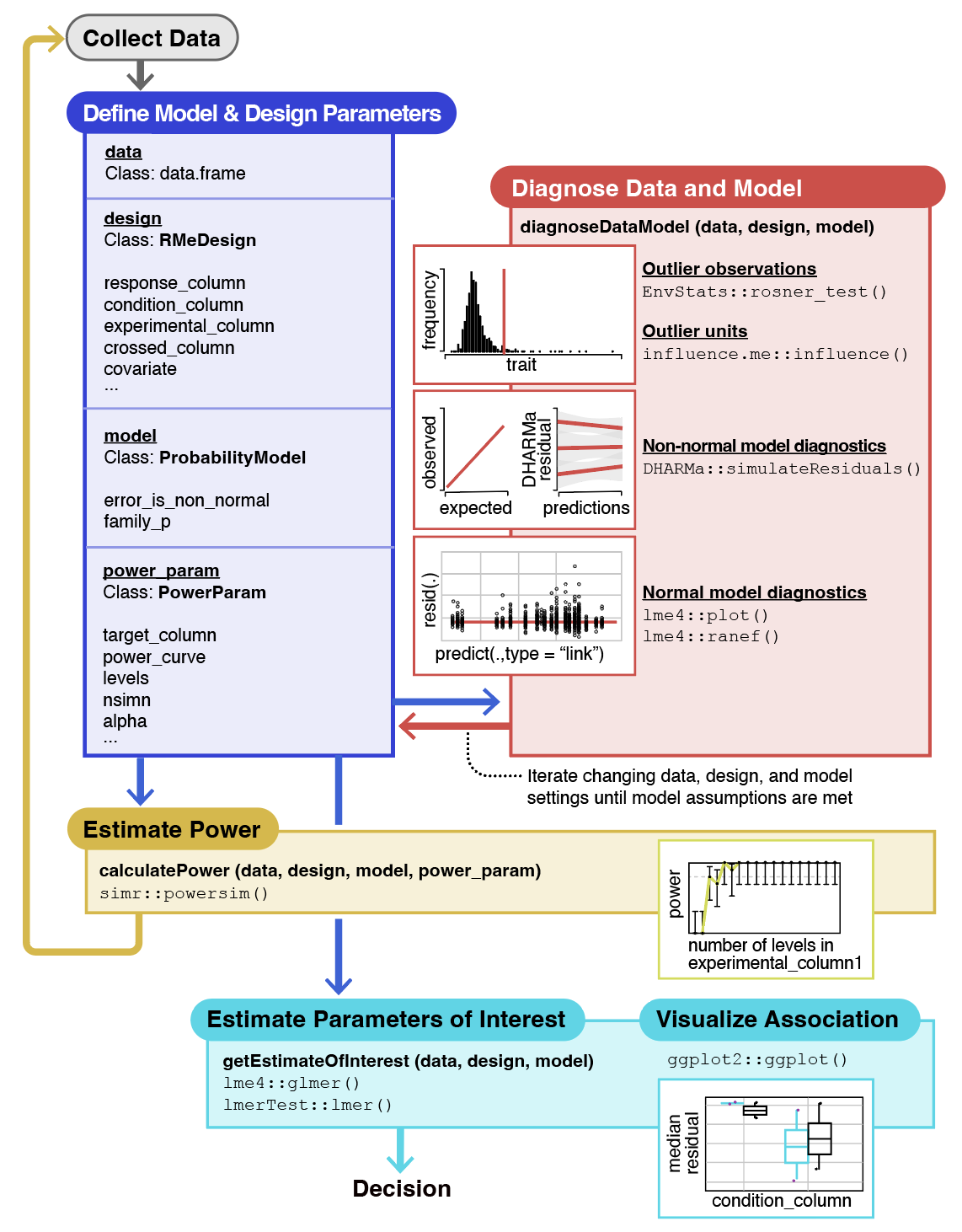
The RMeDPower2 package structure showing the three main S4 classes (RMeDesign, ProbabilityModel, PowerParams) and their three key functionalities of performing 1. quality control checks for model assumptions, identification of outliers, 2. perform sample-size calculations and 3. to derive and visualize estimates of interest in repeated measures experiments. The functionality is adapted from multiple available R packages
Core Components
- RMeDesign: Defines experimental design including response variables, conditions, experimental hierarchy, covariates, and interaction terms
-
ProbabilityModel: Specifies error distribution assumptions (normal vs non-normal, distribution families)
- PowerParams: Contains parameters for power analysis including target variables, effect sizes, simulation parameters
Key Features
- Experimental Design Support: Handles nested and crossed experimental designs with multiple hierarchy levels
- Power Analysis: Simulation-based power calculations using advanced mixed-effects modeling
- Outlier Detection: Two-level outlier identification at observation and group levels
- Model Validation: Comprehensive assumption testing and diagnostic visualizations
- Flexible Distributions: Support for normal, binomial, Poisson, and negative binomial distributions
- Derive estimates of interest: Derive and visualize estimates of interest
Getting Started
- Quick Start Guide - Learn the basics of RMeDPower2
- Complete Tutorial - Comprehensive tutorial with real-world examples
- Configuration Guide - Guide for choosing parameters for different classes
- Function Reference - Complete documentation for all functions